Plant Phylogenetics Lab
Department of Biology , Ball State University
In our lab, we construct evolutionary trees of living organisms, mainly plants, known as phylogenies. We utilize DNA and morphological trait data to build these phylogenies. By analyzing these phylogenies, we investigate how, where, and when different groups of plants (and animals) have evolved over macroevolutionary timescales. Additionally, we contribute to organizing the vast diversity of living organisms into distinct groups based on their evolutionary relationships, a field known as systematics.
Macroevolutionary Dynamics and Speciation in Tropical Plants
The extraordinary diversity of angiosperms worldwide reflects macroevolutionary processes unfolding over deep time, generating both broad distributions and extreme endemism. These forces, most intense in tropical mountains and islands, generate novel traits, reproductive strategies, and ultimately new species. Our lab uses the pantropical tribe Spermacoceae (~1,500 spp.) as a model system to investigate how trait evolution, breeding-system shifts, and genomic change interact to generate biodiversity. We integrate field, phylogenetic, and genomic approaches to reveal how microevolutionary mechanisms scale up to macroevolutionary patterns.

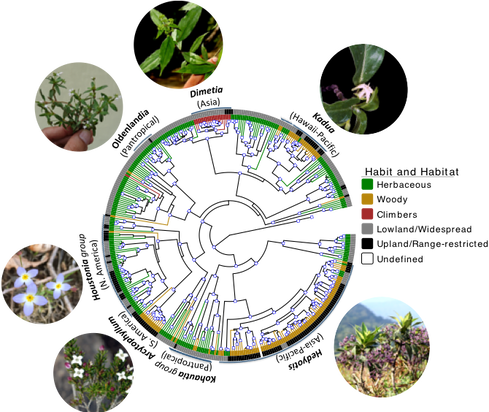
Trait Lability, Polyploidy, and the Genomics of Speciation
- Breeding-system diversity. Heterostyly occurs without clear phylogenetic signal: Houstonia caerulea is distylous (heterostylous), whereas its sister species H. pusilla is homostylous and largely self-fertile. Island radiations show even more extreme shifts—"leaky dioecy" in Hawaiian Kadua and South-Atlantic Nesohedyotis.
- Growth-form transitions. Although ancestrally herbaceous, Spermacoceae has independently evolved woodiness multiple times—especially in Hedyotis, Kadua, and Arcytophyllum. Neupane et al. (2017) link these woody shifts to colonization of montane and insular habitats, where woody forms often dominate.
- Morphological lability. Frequent changes in fruit dehiscence, seed morphology, and pollen traits blur taxonomic boundaries and complicate genus-level delimitation.
- Polyploidy & hybridization. Polyploidy is pervasive, particularly in eastern North-American Houstonia. Church & Taylor (2005) showed genome duplication breaks down diploid reproductive barriers, enabling hybridization; Glennon & Church (2015) found that tetraploids harbor greater genetic diversity and exchange genes largely with other tetraploids. Together, breeding-system shifts, morphological flexibility, and polyploidy forge a dynamic mosaic of reproductive barriers.
Hybridization can either homogenize lineages through ongoing gene flow or, via adaptive introgression and allopolyploidy, accelerate divergence and spawn novel hybrid taxa. Which outcome prevails depends on selection pressures, recombination patterns, and the genomic architecture of barrier loci, all of which vary across landscapes and through time.
Our Current Research Questions
- Geographic patterns of trait evolution: How do breeding-system transitions (heterostyly, homostyly, dioecy), hybridization, and polyploidy differ between island and continental Spermacoceae?
- Polyploid evolution in the Houstonia–Stenaria complex: What is the frequency and evolutionary impact of polyploidy and hybridization events? Do multiple, independent polyploidization events contribute to genetic diversity?
- Genomic mechanisms of speciation: In hyperdiverse, narrowly endemic clades like Hedyotis (~180 spp.) and Kadua (29 spp.), is speciation driven mainly by adaptive radiation in insular or montane settings, or by reticulate processes (hybridization and polyploidy) as in Houstonia? Do genomic signatures point to reinforcement, adaptive introgression, or hybrid speciation as the dominant processes?
- Evolution of woodiness: What genomic changes underlie repeated herbaceous-to-woody transitions in montane Hedyotis, and how do they correlate with diversification?
- Macroevolutionary correlates: Which traits, such as derived woodiness, breeding-system diversity, or polyploidy, best predict elevated diversification rates across the tribe?
By combining fieldwork, phylogenomics, population genomics, comparative transcriptomics, and macroevolutionary modeling, we aim to unravel how micro- and macroevolutionary processes interact to shape the spectacular diversity of tropical, insular, and continental plant lineages.
Taxonomic challenges in Spermacoceae
The high degree of morphological polymorphism and homoplasy in Spermacoceae has made generic delimitation particularly challenging. Historically, taxonomic confusion has arisen from inconsistent and overlapping morphological traits, leading to long-standing disagreements dating back to Linnaeus, who first described many of the group's genera, including Diodia, Hedyotis, Houstonia, Oldenlandia, Richardia, and Spermacoce. Phylogenies of the tribe have revealed that several large genera such as Borreria, Diodia, Hedyotis, Oldenlandia, and Spermacoce, are polyphyletic (Kårehed et al. 2008; Groeninckx et al. 2009). Consequently, many of these genera have been split into monophyletic groups, while others (e.g., Borreria, Oldenlandia, Spermacoce) remain unresolved. My lab is actively addressing these taxonomic problems in collaboration with taxonomists from Argentina and Belgium.
Phylogenomic Congruence
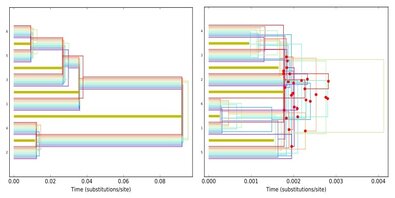
Finding a reliable phylogenetic tree can be challenging due to incongruence between data subsets resulting from stochastic error (e.g. sampling artifacts) or systematic error (e.g. deep coalescence and incomplete lineage sorting that is not accommodated by the model). With the availability of large-scale genomic DNA data from high-throughput sequencing methods, the issue of incongruence becomes even more severe in phylogeny reconstruction. Although some efficient and promising species tree methods are forthcoming, it is always advisable to assess the discordance beforehand, if any, and identify its nature (stochastic or biological) to make a meaningful interpretation of estimated phylogeny from a multi-gene phylogeny. Currently, I am investigating the issues of DNA data combinability (e.g. of different gene regions from targeted sequencing, or next-gen methods), phylogenetic information content, effect of taxonomic sampling and missing data, and the resulting systematic and stochastic bias in the phylogenetic inference under a Bayesian framework. I am particularly exploring the behavior of Bayes Factor (computed using marginal likelihood values) in detecting phylogenomic congruence (alternatively discordance) using simulations designed to create a spectrum of data sets ranging from low to high phylogenetic information content and from complete topological concordance to extreme discordance (due to deep coalescence and subsequent incomplete lineage sorting).
Publications
For the full list of my publications, please visit my Google Scholar profile.
Codes & Scripts
All my scripts, phylogenetic analysis pipelines are available on my GitHub account.
NeupaneLab GitHubLab Members
Ainsley Tierney
Ainsley is a junior biology major with a botany concentration. Her interests include DNA extraction, plant genomics, and plant extremophiles.
Blaire Hayes
Blaire is a junior botany major interested in learning computational techniques for plant phylogenetics.
Greg Monzel
Greg is a B.S./M.S. student investigating the population genomics of American sweet flag (Acorus americanus).
Kieran Claypool
Kieran is a second year MS student. Kieran is working on phylogenomics of North American Spermacoceae and angiosperm 353 phylogeny of Spermacoceae.
Maddie Grove
Maddie is a junior majoring in zoology and botany who loves all things evolution. She assists with DNA isolation and library preparation for Spermacoceae projects.
Nusrat Huda

Nusrat is an M.S. student broadly interested in plant systematics, phylogenetics, and biodiversity. Her research focuses on how plant lineages diversify, particularly within tropical flora. She explores evolutionary questions using both morphological traits and molecular tools, and applies bioinformatics approaches to analyze genetic data and construct phylogenies.
Peyton Rodgers
Peyton is a senior botany major with interests in applied plant sciences. She is currently conducting chromosome counts and ploidy determination in North American Houstonia.
Curriculum Vitae
Download my CV here.
Contact Information
Dr. Suman Neupane
Department of Biology
Ball State University, Muncie, IN 47306
Email: suman [dot] neupane [at] bsu [dot] edu
Tutorials
- Introduction to Phylogenetic Analysis
- Comparative Methods in R
- Plant Specimen Preservation